The key research questions of the lab are:
- How do we engineer wearable sensor systems for the real world?
- What new machine learning/AI models should we build to understand signals from the body?
- Where are the untapped opportunities in clinical medicine to better understand individual physiology?
The Problem

The vast majority of patients spend a relatively small fraction of their total time in a clinic. Most of our time is spent on the rest of life, which includes factors like diet and sleep that play a huge role in our health, but remain mostly out of the purview of the current healthcare system.
This excludes a lot of valuable information. What about….
- … everything we experience outside of the clinic?
- … situations where patients cannot speak for themselves?
- … functions outside of our conscious awareness and control?
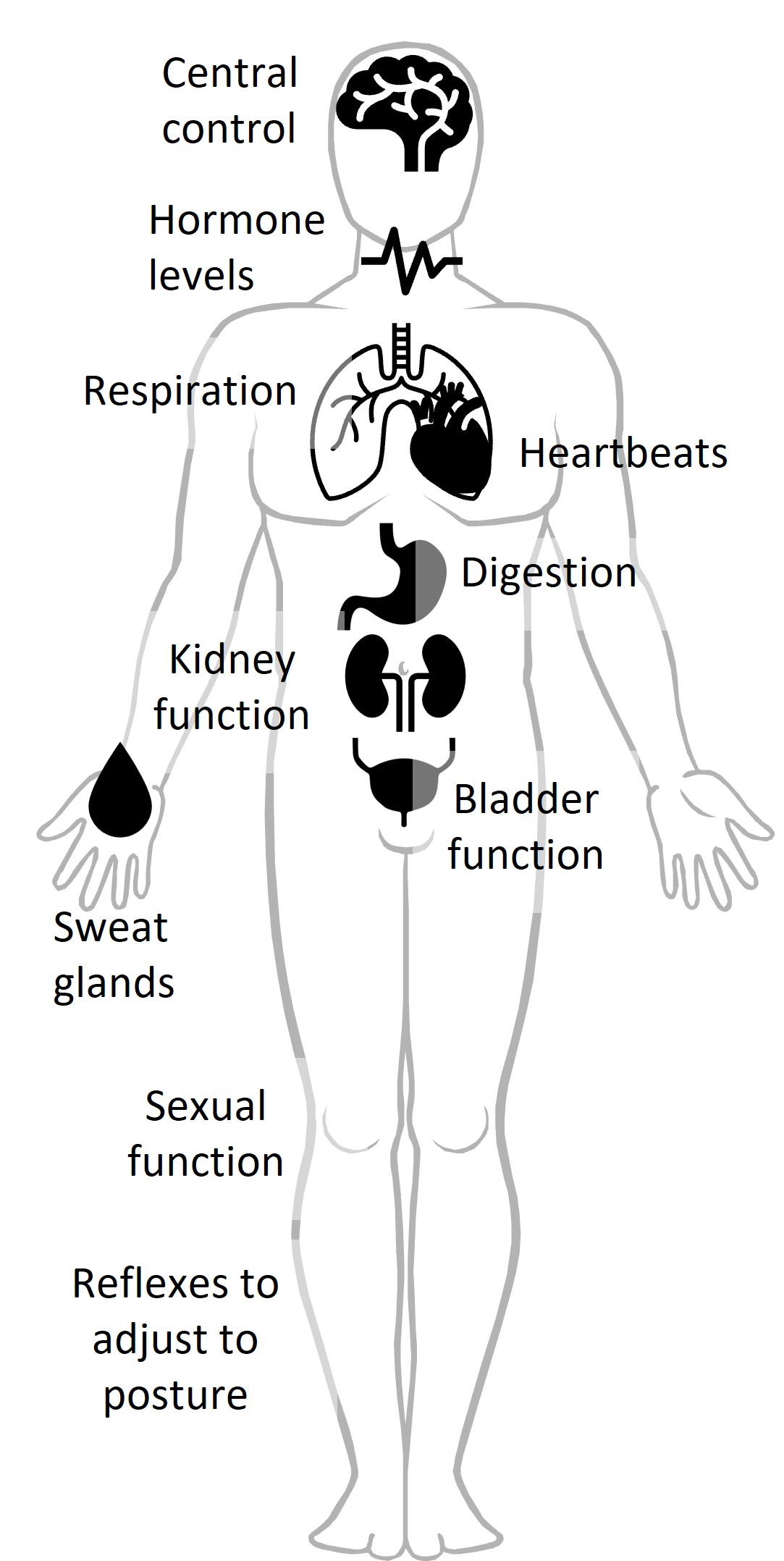
The Autonomic Nervous System
One answer to addressing these challenges is taking advantage of the autonomic nervous system. The autonomic nervous system controls all of our unconscious function (e.g. heartbeats, breathing, digestion, sweating, hormone production, etc.) Advances in wearable sensing over the last decade allow us to track it in ways we could not before. This means it can be monitored:
- outside of the clinic
- even when people are unconscious or incapacitated
- to capture body function beyond conscious control
Our Interdisciplinary Approach
We take a highly interdisciplinary approach to address the main areas of need: new modalities of information, new models and methods, and new applications. Our work falls into three parallel and synergistic lines of inquiry: clinically immersive data collection and system prototyping, algorithm development, and physiologic modeling.
Clinically immersive data collection and system prototyping
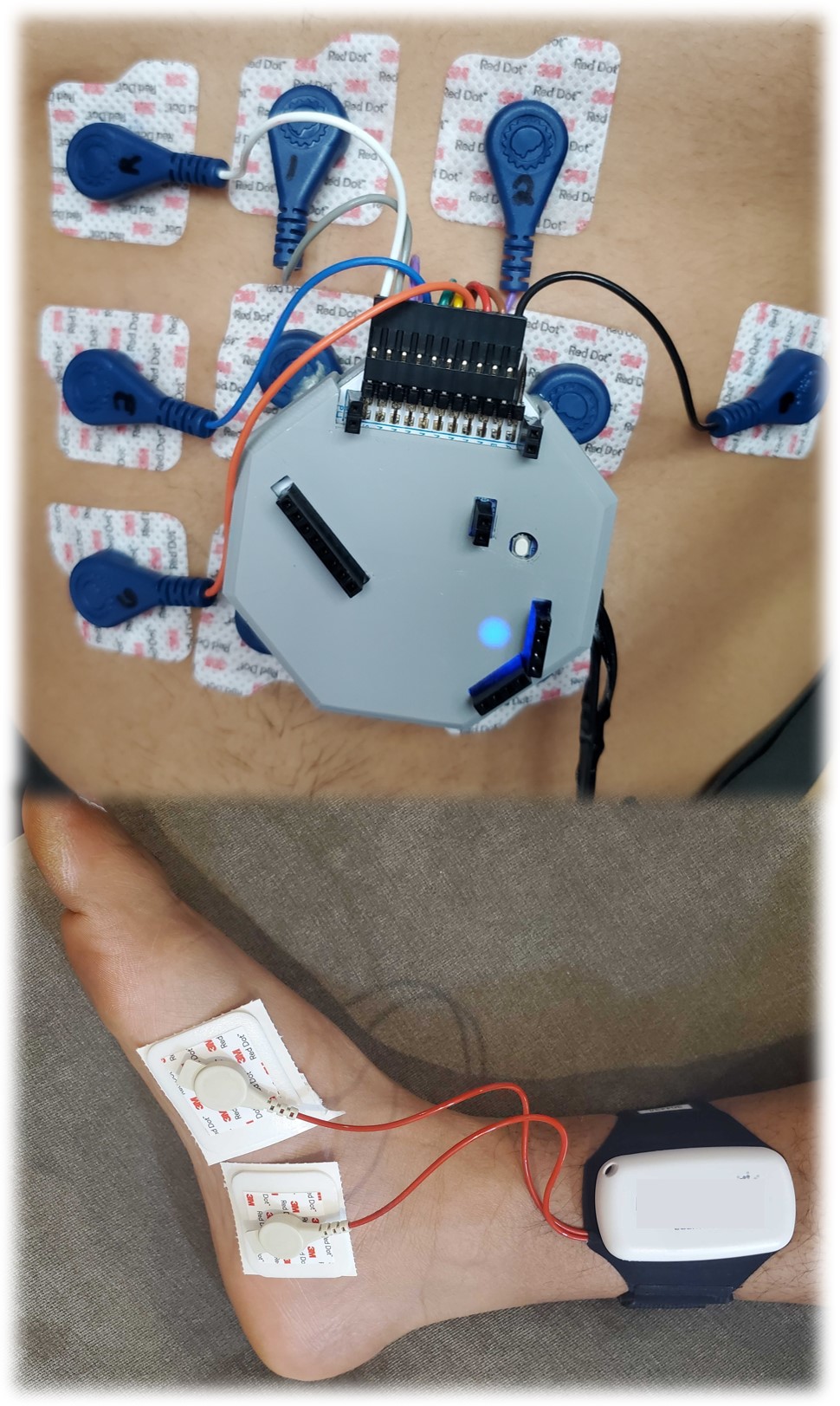
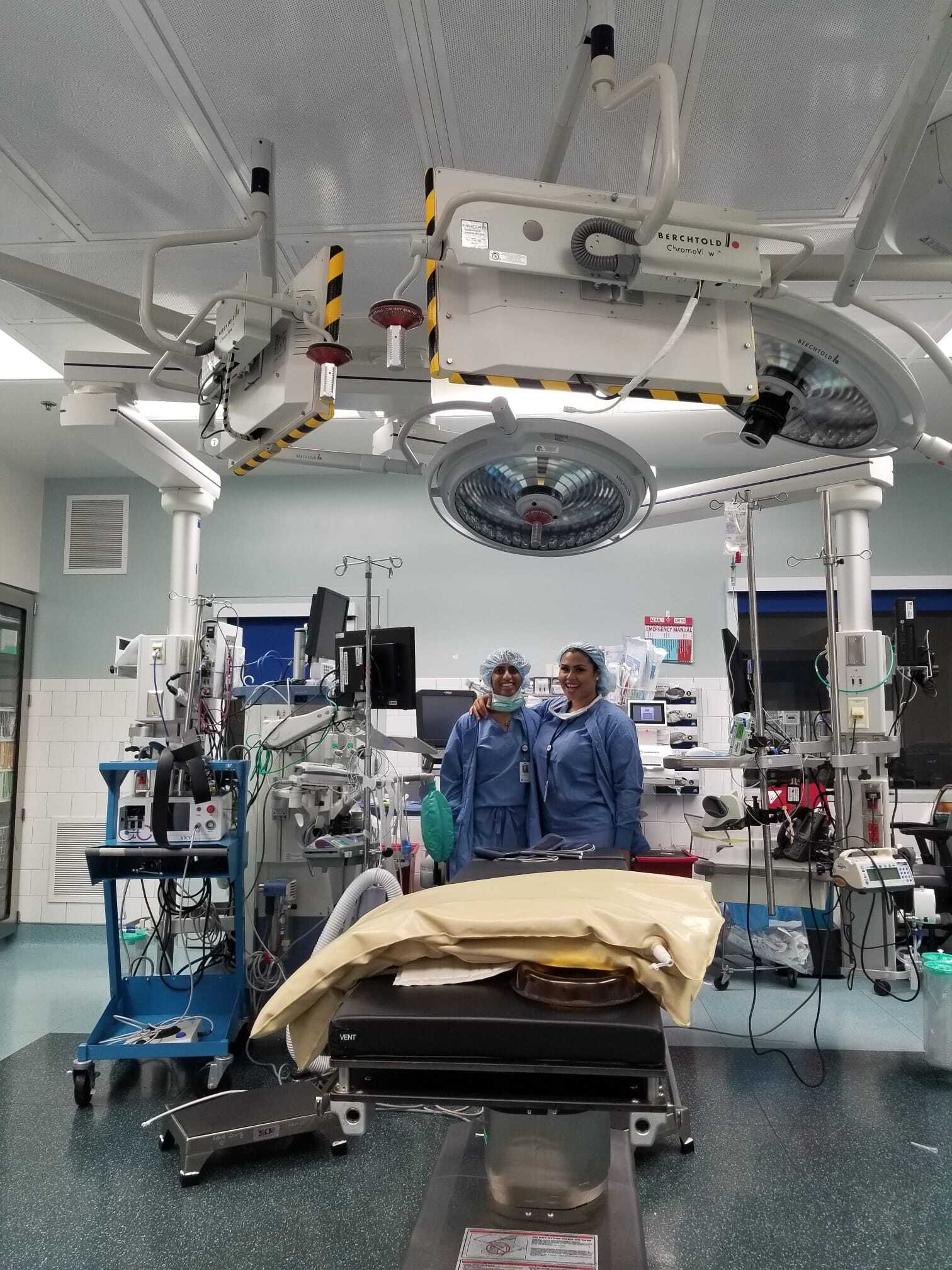
We collect our own data by running human studies with healthy volunteers and patient populations, both in clinical settings and at home. Each of these data collection scenarios presents unique challenges that require creative solutions from the hardware standpoint. Our goal is to balance (1) the need for high-quality clinical-grade data with (2) time and resource constraints of busy clinics and clinicians and (3) usability and convenience for the patient. This is an area we hope to expand with more hardware prototyping in the future!
Previous human studies:
- Modulation of autonomic activity in a controlled setting in healthy volunteers
- Monitoring multi-sensor activity in patients undergoing surgery while under anesthesia
- Tracking 24-72 hours of continuous multi-sensor autonomic activity at home in healthy volunteers and patients with chronic migraine
Current/ongoing work:
- Developing new wearables for the tracking bladder muscle activity at home in multiple sclerosis
- Developing v2 of our full at-home multi-sensor autonomic platform that is energy-efficient and easy to use
Algorithm development
Collecting data in challenging real-world settings (in and out of the clinic) results in similarly challenging data. These data are noisy, filled with known and unknown artifacts, incomplete, and either lacking ground truth or labeled with annotations that are often inexact or even incorrect due to human error. Algorithm development, especially focused on robust, scalable automation without supervised (labeled datasets) is a major challenge. These methods must learn complex patterns that may be hard to see without losing physiologic integrity of the data (e.g. the point process nature of underlying signals). There are many opportunities to develop neural architectures and differentiable algorithms for biosignals for both single sensor and multimodal data.
Examples of previous work:
- Quantifying the physiology of sweat gland activity from first principles and biophysics (IEEE TBME 2021, PLoS Comp Bio 2021, PNAS 2020, Proc IEEE EMBC 2019, Proc IEEE EMBC 2018)
- Automatic artifact removal from electrodermal activity data in the operating room (Phys Meas 2022, Proc IEEE EMBC 2021)
- Automated classification of sleep and wake from triaxial accelerometer data (Proc IEEE EMBC 2022)
- Neural temporal point process methods for modeling heartbeat dynamics (ICLR TS4H Workshop 2024)
Current/Ongoing work:
- Network models of the ANS across the body
Physiologic modeling
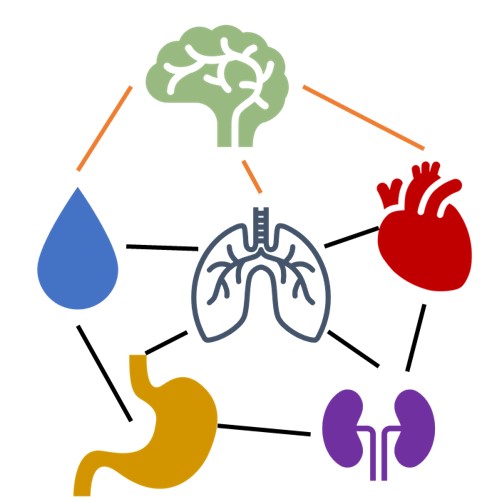
At the core of all of these exciting new applications is finding new and creative ways to mathematically model what we know of underlying physiology, especially at the macroscale/organismal level. For example, how does the ANS act as a dynamic network across different organ systems? We aim to build physiologically and statistically rigorous models that are also interpretable for scientists and clinicians. One of our interests is embedding rich inductive biases drawn from physiological priors into computational models to enhance their performance even on small datasets.
Examples of previous work:
- Using autonomic responses to monitor general anesthesia (PLoS ONE 2021, Proc IEEE EMBC 2022, Proc IEEE EMBC 2020, Proc IEEE ESGCO 2020, Proc CinC 2020)
- Tracking 24-hour autonomic activity at home in different types of gastroparesis (IEEE TBME 2023, Proc IEEE EMBC 2023)
- EEG-based patient stratification in chronic pain (Proc IEEE EMBS Data Sci)
- Tracking unconscious pain during surgery (PNAS)
Current/Ongoing work:
- Characterizing multi-system ANS activity in long COVID and chronic fatigue syndrome
- Piloting the at-home autonomic clinic of tomorrow